Minc Lab | Research
Cells come with stereotypical shapes and sizes which are often associated with their function. Neurons grow in highly elongated morphologies to build neuronal networks, while red blood cells have discoid-like deformable morphologies to squeeze within tiny blood vessels. Questions that fascinate our team are to understand how cells define their particular shape, and how information in cell geometry may be conveyed into cell function or in organizing the cell interior. To address these fundamental questions, we integrate original methods at the frontier between molecular biology, genetics and physics and mathematics. One of our ultimate goal is to capture biological behavior with mathematical models.
Minc Lab | Research
Cells come with stereotypical shapes and sizes which are often associated with their function. Neurons grow in highly elongated morphologies to build neuronal networks, while red blood cells have discoid-like deformable morphologies to squeeze within tiny blood vessels. Questions that fascinate our team are to understand how cells define their particular shape, and how information in cell geometry may be conveyed into cell function or in organizing the cell interior. To address these fundamental questions, we integrate original methods at the frontier between molecular biology, genetics and physics and mathematics. One of our ultimate goal is to capture biological behavior with mathematical models.
Minc Lab | Research
Cells come with stereotypical shapes and sizes which are often associated with their function. Neurons grow in highly elongated morphologies to build neuronal networks, while red blood cells have discoid-like deformable morphologies to squeeze within tiny blood vessels. Questions that fascinate our team are to understand how cells define their particular shape, and how information in cell geometry may be conveyed into cell function or in organizing the cell interior. To address these fundamental questions, we integrate original methods at the frontier between molecular biology, genetics and physics and mathematics. One of our ultimate goal is to capture biological behavior with mathematical models.
Division Plane Positioning
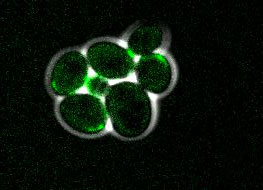
The proper positioning of the site of cell division is crucial not only for the survival of all cells, but also for the development of multi-cellular tissues. The lab is fascinated by the process of early embryogenesis during which the egg becomes fertilized and follows a stereotypical choreography of successive division, called cleavage patterns. Our goal is to understand, predict and establish quantitative laws that explain the patterning of cell divisions in early embryos. To this end, we combine mathematical modelling, imaging and biophysics approaches. Our model experimental system is the sea urchin embryo, which integrates many advantages for the study of early development. To extend our findings to other multicellular context, we also have collaboration with other labs, working on flies, frogs, fishes and ascidians.
video: Dividing sea urchin embryos
Sea urchin embryos going through early development. One can observe the centration and fusion of nuclei and the successive division events which are typical of early embryos. Time is min:sec. (Credit: Nicolas Minc)
Jing Xie; Jeremy Sallé; Aude Nommick; Luc Lederer; Serge Dmitrieff; Javad Najafì; Nicolas Minc
Collaborations:
- Guillaume Romet-Lemonne (IJM, Paris)
- Aki Kimura (NIG, Japan)
- Yohanns Bellaiche (Institut Curie, Paris)
- Delphine Delacour (IJM, paris)
- Xie J, Najafi J, Le Borgne R, Verbavatz JM, Durieu C, Sallé J, Minc N. (2022) “Contribution of cytoplasm viscoelastic properties to mitotic spindle positioning”
Proc Natl Acad Sci U S A. 22;119(8):e2115593119. - Palenzuela H, Lacroix B, Sallé J, Minami K, Shima T, Jegou A, Romet-Lemonne G, Minc N. (2020) “In Vitro Reconstitution of Dynein Force Exertion in a Bulk Viscous Medium.”
Curr Biol. 30(22):4534-4540.e7. - Tanimoto H, Sallé J, Dodin L, Minc N. (2018) “Physical Forces Determining the Persistency and Centering Precision of Microtubule Asters.”
Nat Phys. 2018 Aug;14(8):848-854. - Pierre A, Sallé J, Wühr M, Minc N. (2016) “Generic Theoretical Models to Predict Division Patterns of Cleaving Embryos.”
Developmental Cell 39(6):667-682 - Bosveld F, Markova O, Guirao B, Martin C, Wang Z, Pierre A, Balakireva M, Gaugue I, Ainslie A, Christophorou N, Lubensky DK, Minc N, Bellaïche Y, (2016) “Epithelial tricellular junctions act as interphase cell shape sensors to orient mitosis” Nature 530, 495–498
- Minc N. , Burgess, D. and Chang F. (2011), “Influence of cell geometry on division plane positioning” Cell., 144 (3): 414-426.



Cell shape and polarity
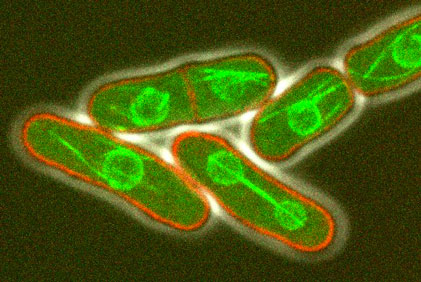
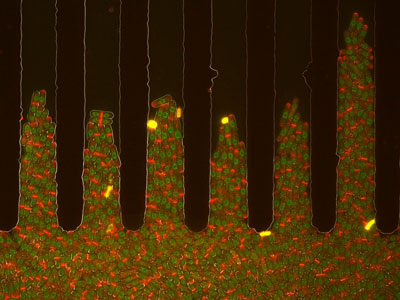
The morphology of single cells as well as cells embedded in tissues underlie many of their function. We seek to address how cells develop their particular shape and internal organization. For instance, many cells only grow at a given location, a process called polarized growth, which involves specific biochemical reactions needed to cluster a local domain of growth activity. To study these aspects we use a unicellular fungi called fission yeast. These cells have rod-shapes and are genetically tractable, with many mutants defective in morphogenesis. These cells are encased in a stiff cell wall and are turgid, which provides them with particular mechanical properties. One of our ultimate goal is to link biochemical and biomechanical signals which serve to shape cells.
video: Growing and dividing fission yeast cells
Fission yeast cells labeled for the nucleus in red and for a polarity marker (bgs4, a cell wall enzyme) in green grow and divide along several cell cycles. Time is in hs:min. (Credit: Nicolas Minc)
Fatima Hamdi; Celia Municio Diaz; Louis Chevalier; Nicolas Minc
Collaborations:
- Etienne Couturier (MsC, Paris)
- Arezki Boudaoud (ENS Lyon)
- Miguel Peñalva (U. madrid)
- Yolanda Sanchez (U. Salamanca)
- Neeli-Venkata R, Diaz CM, Celador R, Sanchez Y, Minc N. (2021) “Detection of surface forces by the cell-wall mechanosensor Wsc1 in yeast.”
Dev Cell. ;56(20):2856-2870.e7. - Davì V, Chevalier L, Guo H, Tanimoto H, Barrett K, Couturier E, Boudaoud A, Minc N. (2019) “Systematic mapping of cell wall mechanics in the regulation of cell morphogenesis.”
Proc Natl Acad Sci U S A. ;116(28):13833-13838. - Haupt A, Ershov D, Minc N. (2018) “A Positive Feedback between Growth and Polarity Provides Directional Persistency and Flexibility to the Process of Tip Growth.” Curr Biol. ;28(20):3342-3351.e3.
- Davì V, Tanimoto H, Ershov D, Haupt A, De Belly H, Le Borgne R, Couturier E, Boudaoud A, Minc N. “Mechanosensation Dynamically Coordinates Polar Growth and Cell Wall Assembly to Promote Cell Survival.” Dev Cell. 2018 Apr 23;45(2):170-182.e7.
- Bonazzi D, Julien JD, Seddiki R, Romao M, Piel M, Boudaoud A, Minc N, (2014) “Symmetry breaking in spore germination relies on an interplay between polar cap stability and spore wall mechanics.” Developmental Cell, 28 (5):534-546



MORE VIDEOS
Minc Lab | Protocols and scripts
Models
Division predict
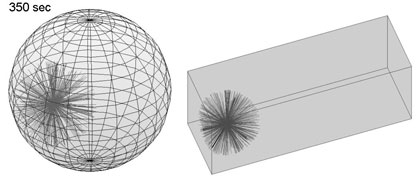
This simple Matlab script, developped by Nicolas Minc (Minc et al. Cell 2011; Campinho et al NCB, 2013), serves to predict the direction of the cell division axis based on cell shape. It interacts with the user and uses hand contour tracing to infer cell geometry and outputs an energy landscape for spindle orientation angles.
3D Model for Cleavage Patterns
This package, developped by Dmitry Ershov and Anaëlle Pierre ( Ershov and Minc, Methods Mol Biol, 2019; Pierre et al. Dev Cell 2016), includes Matlab files, and surface evolver guidelines to predict cell division axis, in 3D , based on multiple inputs (cell shape, polarity, yolk gradients etc...). for the cleavage patterns of early embryos, or in any cellular or tissular context.
Image analysis
WallTracker
This Matlab package, developped by Hirokazu Tanimoto, Dmitry Ershov and Valeria Davi (Davi et al. Dev Cell 2018), serves to compute cell wall thickness in live fission yeast cells from the fluoresence of a membrane marker and a lectin marking the outer part of the wall.
WallTracker 2.0
This Matlab package, developped by Hirokazu Tanimoto and Valeria Davi (Davi et al. In revision 2019), is a second version of Wall tracker with a specific module to analyse shape changes following cell wall ablation; which serves to compute cell wall elasticity and thickness in fission yeast and other walled cells.
Pombenator 2.6
This Matlab package, developped by Dmitry Ershov (Taheraly, Ershov et al. J. Cell Science 2020), allows to compute single tip growth as well as the concentration of fluorescent protein at different positions in cells such as fission yeast and filamentous fungi.
CurvPol
This Matlab package, developped by Hirokazu Tanimoto (Bonazzi, Haupt, Tanimoto .Curr Biol 2015), serves to quantify polarity domain size and local cell curvature at the polarity site. It interacts with the user and uses hand contour tracing to obtain a detailed measurment of relevant parameters.
PombeSizer
This simple Matlab script, developped by Yonatan Zegman and Nicolas Minc (Zegman et al. Methods Cell Biology 2015), serves to quantify size parameter of single pombe cells. It interacts with the user and uses hand contour tracing to obtain a detailed measurement of cell volume, surface and other parameters.
